Nucleic Acid Purification Kits
Nucleic acid purification kits are used to extract and purify nucleic acids (DNA or RNA) from your samples, including cells, tissues, etc. DNA or RNA is an organic molecule that store genetic information in the cells. To study them, we have to isolate and purify the nucleic acids from the unwanted component in the cells. But before that, let’s have a look at what are the nucleic acids.
What are the nucleic acids?
Living organisms on this earth, either unicellular or multicellular organisms, contain two major types of nucleic acids. They are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).
Both of them made from nucleotides, organic molecules that contain five-carbon sugar backbones, phosphate groups and nitrogen bases.
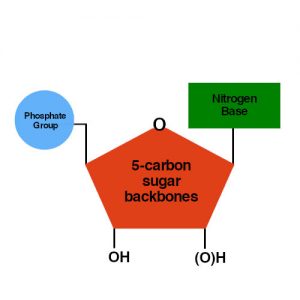
A nucleic acid backbone consists of 1 nitrogen base, 1 carbon sugar backbone and 1 phosphate group. RNA structure contains (OH) group in the 2′ while DNA only contains (H) group.
Deoxyribonucleic Acid (DNA)
DNA is a polymer of 4 nucleotides, Adenine (A), Thymine (T), Cytosine (C) and Guanine (G), where A pair with T and C pair with G. They contain all the genetic code that is essential for development and function of every single cell. Besides, they are the fundamental molecules that make us different from each other!
Ribonucleic Acid (RNA)
Similar to DNA, RNA is also a polymer of 4 nucleotides, Adenine (A), Uracil (U), Cytosine (C) and Guanine (G). Instead of Thymine as in DNA, RNA contains Uracil (U), which also pair with Adenine (A).
Up to date, there are three main types of RNA:
- Messenger RNA (mRNA) – its primary role is to encode the amino acid sequence of polypeptides, which eventually become part of a protein.
- Ribosomal RNA (rRNA) – it is part of the ribosomes, which responsible for translation, in other words, synthesis of protein.
- Transfer RNA (tRNA) – it helps to bring amino acid to the ribosome and decode the mRNA sequence into protein.
Why need to purify DNA and RNA from your samples?
DNA or RNA purifications are the fundamental steps for any downstream application in biotechnology and molecular biology study. Without purifying your samples, it may contain many contaminants that will lead to erroneous result in your research.
Hence, using DNA or RNA isolation kit can help us to:
- Obtain high quality of DNA and RNA from your sample
- Reduce the amounts of contaminants which may shorten the shelf life of your DNA and RNA
For different applications, we need to use different kinds of purification kits.
Monarch Plasmid Miniprep Kit
Plasmid is a small circular DNA molecule that physically separated from chromosomal DNA. They commonly found in bacterial cells, and sometimes in archaea and eukaryotic organisms. They can replicate independently and often carry genes that benefit the cells survival. One of the common genes is the antibiotic resistance gene.
Scientists usually used plasmid for transformation, transfection, DNA sequencing, restriction digestion and other enzymatic manipulations. In order to do so, the high quality of purified plasmid DNA is a must!
The Monarch Plasmid Miniprep Kit is a fast and reliable kit to purify the high quality of plasmid DNA. Here, it employs standard cell resuspension, alkaline lysis, and neutralization steps. To remove all the unwanted substances, like salts, proteins, RNA and other cellular components, NEB uses a unique washing buffer to do so, leaving low-volume elution of concentrated and highly pure DNA. Besides, NEB included colour indicators to ease users in monitoring the completion of extraction.
What do you need to know before using Monarch Plasmid Miniprep Kit?
Here, we show you the specification in the below table:
| Description | Specifications |
| Culture Volume | Recommended between 1 to 5 ml and not to exceed 15 OD units |
| Binding Capacity | Up to 20μg |
| Plasmid Size | Up to 20kb |
| Typical Recovery | Up to 20μg. Yield is affected by several factors, including plasmid copy number, host strain, culture volume, and growth conditions |
| Elution Volume | Elute in as little as 30μL |
| Purity | A260/280 and A260/230 ⥠1.8 |
| Protocol Time | Spin and incubation times: 10.5 mins |
| Downstream Application | Transformation, Transfection, DNA sequencing, Polymerase Chain Reaction (PCR), Labelling, Restriction Digestion and Other Enzymatic Manipulations, etc. |
Advantages of using Monarch Plasmid Miniprep Kit
- Low elution volume: as little as 30μL
- Optimized column design to prevent buffer retention and salt carry-over – say no more to contamination!
- Faster protocols and less spin time – saving you valuable time
- Easily keep track of the certain steps by observing the colour changes of buffer
- No RNase needed before starting – save money
- Easily label columns – tab and frosted surface
- Buffers and columns can be purchased separately
- Go GREEN – significantly less plastic is used (compare to other kits) and recyclable packaging
- No hazardous materials fees – save money and save the environment!
Basic Steps for Plasmid Purification using Monarch Plasmid Miniprep Kit
**Before you start, add 4 volumes of ethanol to 1 volume of DNA wash buffer.
**All centrifugation steps should be carried out at 16,000 x g (~13,000 RPM).
Pelleting
Pellet your cells between 1-5ml of bacterial cell culture by centrifuge at 16000g / 13000 rpm for 30 seconds. Carefully discard the supernatant.
Resuspend Pellet
Resuspend you pellet with 200μl Monarch Plasmid Resuspension Buffer (B1, Pink in colour). Vortex or pipet for thorough mixing until completely resuspended.
Tips: Resuspend cells completely to achieve complete lysis in the following steps.
Lyse Bacteria
Add 200μl of Monarch Plasmid Lysis Buffer (B2, Blue in colour) to the cell suspension. Mix gently by inverting the tubes 5-6 times until the colour changes to dark pink. Incubate for 1 min at room temperature to ensure complete lysis of the cells.
Tips: Do not vortex the solution as this may shear your DNA and the quality and yield of extracted DNA can be affected.
Neutralize Lysate
Add 400μl of Monarch Plasmid Neutralization Buffer (B3, Yellow in colour) to the solution. Gently mix the solution by inverting the tube 4-6 times. Ensure that you mix the solution until uniformly yellow and precipitate forms. Incubate for 2 mins at room temperature and centrifuged for 2-5 mins.
Tips: Longer spin times will result in a more compact pellet, which will lower the risk of clogging the column.
Binding DNA to Column
Transfer the supernatant which contains the plasmid DNA to the spin column and centrifuge for one min. The DNA is now bound to the column matrix. Remove the column and discard the flow-through.
Washing Steps
Insert column into the collection tube and add 200μl of Monarch Plasmid Wash Buffer 1. Centrifuge for 1 min. Optional: You may discard the flow-through. Add 400ul Monarch Plasmid Wash Buffer 2 and centrifuge for 1 min. Transfer the column to the clean centrifuge tube.
Tips: Ensure the column doesn’t touch flow-through during transfer.
Elution
Add minimum 30ul Monarch DNA Elution Buffer to the centre of the column matrix. Incubate the solution for 1 min at room temperature. Centrifuge 1 min and collect the flow-through and your purified plasmid DNA is ready for any of your downstream application.
Tips: Incubation less than 1 min can reduce the yield of DNA.
For video, please watch Protocol for Monarch Plasmid Miniprep Kit.
How to maximize yield when using Monarch Plasmid Miniprep Kits?
Here are some tips to maximize the yield of plasmid DNA.
- Ensure that the pellet is resuspended completely
- Bacteria pellet that is not resuspended completely won’t be lysed efficiently and the yield of DNA after purification is lower.
- Ensure the solution is mixed completely by inverting the tubes gently during cell lysis.
- Make sure you add the elution buffer to the centre of the matrix without puncturing it. This will increase the efficiency of elution.
- Be sure to incubate the elution buffer in the column for a full minute before spinning.
Monarch PCR & DNA Cleanup Kit
After PCR or other enzymatic reactions, you might need to purify the DNA, for use in restriction digests, DNA sequencing, ligation or other enzymatic manipulations. With Monarch PCR & DNA Cleanup kit, you can easily and quickly purify the high quality of DNA.
This kit utilizes a bind/wash/elute workflow with minimal incubation and spin times. Besides DNA, the protocol can be modified to purify smaller DNA fragments, including oligonucleotides and ssDNA.
What do you need to know about Monarch PCR & DNA Cleanup Kit?
| Description | Specifications |
| DNA Sample Type |
|
| Binding Capacity | Up to 5μg |
| DNA Size Range |
|
| Typical Recovery |
|
| Elution Volume | ⥠6 μl |
| Purity | A260/280 > 1.8 and A260/230 > 1.8 |
| Protocol Time | 5 minutes of spin and incubation time |
| Downstream Application | Ligation, restriction digestion, labelling and other enzymatic manipulations, library construction and DNA sequencing. |
In what applications you should use Monarch PCR & DNA Cleanup Kit?
| Application | Explanation |
| PCR cleanup | Purify your DNA by removing the polymerases, primers, detergent, etc. that used in PCR amplification. |
| Enzymatic Reaction Cleanup | Remove the restriction enzymes and modifying enzymes (ligases, kinases, nucleases, etc) and allowing for effective desalting and concentration of your DNA sample. |
| cDNA Cleanup | Purify your cDNA by removing RT nd polymerase as well as nucleotides. |
| Labelling Cleanup | Remove unincorporated radiolabeled or fluorescently labelled nucleotides from your DNA. |
| Plasmid Cleanup | Plasmid from unknown sources may contain inhibitors and unwanted contaminants. You can remove these unwanted substances using this kit. |
Advantages of using Monarch PCR & DNA Cleanup kit
- Low elution volume: elute in as little as 6 μl
- Prevent buffer retention and salt carry – over with optimized column design – say no more to contamination!
- Save time with fast, user-friendly protocols
- No need to monitor pH, as NEB already optimized the buffer solution for you
- Protocol modification allows for ssDNA purification, oligonucleotide purification, and purification of other small DNA fragments
- Buffers and columns available separately – which means you can buy either buffers or columns only if you have finished one of them
- GO GREEN! – Significantly less plastic used when compared with other kits and responsibly-sourced and recyclable packaging
Monarch PCR and DNA Cleanup Kit Protocols
**Before you start, add 4 volumes of ethanol to 1 volume of DNA wash buffer.
**All centrifugation steps should be carried out at 16,000 x g (~13,000 RPM).
Dilution of Sample
Only use 20-100μL of your DNA sample and dilute with Monarch DNA Cleanup Binding Buffer according to your sample type:
| Description | Specification |
| DNA Sample Type | DNA from PCR or other enzymatic reaction ssDNA or dsDNA oligonucleotides can also be purified with using Oligonucleotide Cleanup Protocol |
| Binding Capacity | Up to 5μg |
| DNA Size Range | ~50 bp to 25 kb DNA ⥠15 bp to 25 kb (dsDNA) and DNA ⥠18 nt to 10 kb (ssDNA) can also be purified using the Oligonucleotide Cleanup Protocol |
| Typical Recovery | DNA (50 bp to 10 kb): 70–90% DNA (11–23 kb): 50–70% ssDNA ⥠18 nt and dsDNA ⥠15 bp: 70-85% |
| Elution Volume | ⥠6 μl |
| Purity | A260/280 > 1.8 and A260/230 > 1.8 |
| Protocol Time | 5 minutes of spin and incubation time |
| Downstream Application | Ligation, restriction digestion, labelling and other enzymatic manipulations, library construction and DNA sequencing. |
When do you need to use Monarch PCR & DNA Cleanup Kit?
| Application | Explanation |
| PCR cleanup | Purify your DNA by removing the polymerases, primers, detergent, etc. that used in PCR amplification. |
| Enzymatic Reaction Cleanup | Remove the restriction enzymes and modifying enzymes (ligases, kinases, nucleases, etc) and allowing for effective desalting and concentration of your DNA sample. |
| cDNA Cleanup | Purify your cDNA by removing RT nd polymerase as well as nucleotides. |
| Labelling Cleanup | Remove unincorporated radiolabeled or fluorescently labelled nucleotides from your DNA. |
| Plasmid Cleanup | Plasmid from unknown sources may contain inhibitors and unwanted contaminants. You can remove these unwanted substances using this kit. |
Advantages of Monarch PCR & DNA Cleanup kit
- Low elution volume: elute in as little as 6 μl
- Prevent buffer retention and salt carry-over with optimized column design – say no more to contamination!
- Save time with fast, user-friendly protocols – save your valuable time
- No need to monitor pH, as NEB already optimized the buffer solution for you
- Protocol modification allows for ssDNA purification, oligonucleotide purification, and purification of other small DNA fragments
- Buffers and columns available separately – which means you can buy either buffers or columns only if you have finished one of them
- GO GREEN! – Significantly less plastic used when compared with other kits and responsibly-sourced and recyclable packaging
Monarch PCR and DNA Cleanup Kit Protocols
**Before you start, add 4 volumes of ethanol to 1 volume of DNA wash buffer.
**All centrifugation steps should be carried out at 16,000 x g (~13,000 RPM).
Dilute Your Sample
Only use 20-100μL of your DNA sample and dilute with Monarch DNA Cleanup Binding Buffer according to your sample type:
| Sample Type | Ratio of Binding Buffer of Sample | Example |
| dsDNA > 2kb (plasmids, gDNA) | 2:1 | 200μl:100μl |
| dsDNA < 2kb (some amplicons, fragments) | 5:1 | 500μl:100μl |
| ssDNA (cDNA, M13) | 7:1 | 700μl:100μl |
Tips: If your initial DNA sample volume is less than 20μL, you need to top up with TE buffer to at least 20μL. However, if your initial DNA sample volume is more than 800μL, you have to load the sample incrementally to avoid overflowing column.
Bind Your DNA to Column
Load your sample to a column seeded to a collection tube and close the gap. Spin for one min and the DNA is bound to the column matrix. Discard the flow-through.
Wash Your DNA
Reinsert the column into the collection tube and add 200μL of DNA Wash Buffer. Spin for one min. Repeat the wash steps by adding 200μL of DNA wash buffer and spin again for one min.
Elute Your DNA
Transfer the column into a clean tube. Add a minimum of 6μL DNA Elution Buffer to the centre of the column and incubate for one min for maximizing yield. Spin for one min and collect the flow-through, your DNA is now ready for any downstream application.
Tips: When transferring the column, make sure that the tip doesn’t touch the flow-through to avoid contamination.
If you want to save time, you can reduce the binding and elution spins to 30 seconds.
For video, please watch Monarch PCR and DNA Cleanup Kits Protocol.
How to maximize DNA yield when using Monarch PCR and DNA Cleanup Kit?
- Add elution buffer to the centre of the column matrix for efficient elution.
- For DNA > 10kb, heat elution buffer to 50°C prior to adding to the column to improve yield.
- Sequential elution may increase yield but will result in lower final DNA concentration
Monarch DNA Gel Extraction Kit
After running DNA gel electrophoresis, we usually want to extract the DNA interest from the agarose gel. By using Monarch DNA Gel Extraction kit, you can rapidly and reliably get up to 5μg of purified, high-quality DNA from agarose gels.
Like other purification methods, Monarch DNA Gel Extraction kit also utilizes a bind/wash/elute method in purifying DNA. On top of that, the column in this kit ensures zero buffer retention and no carryover contaminants, allowing low sample elution volume (6μL).
To make the kit as user-friendly as possible, NEB has optimized the buffers. You do not need to monitor pH during purification of DNA. What’s more exciting is you do not need isopropanol to melt the agarose, saving you a step and time. After elution, the eluted DNA is ready for use in restriction digests, DNA sequencing, ligation and other enzymatic manipulation.
Specifications of Monarch DNA Gel Extraction kit
| Description | Specification |
| DNA Sample Type | double-stranded DNA from agarose gels |
| Binding Capacity | up to 5 μg |
| DNA Size Range | ~50 bp to 25 kb |
| Typical Recovery | DNA (50 bp to 10 kb): 70–90% DNA (11–23 kb): 50–70% |
| Elution Volume | ⥠6 μl |
| Purity | A260/280 > 1.8 |
| Protocol Time | 10 minutes of spin and incubation time |
| Compatible Downstream Application | Ligation, restriction digestion, labelling and other enzymatic manipulations, library construction and DNA sequencing. |
Advantages of using Monarch DNA Gel Extraction Kit
- Elute in as little as 6μl
- Prevent buffer retention and salt carry-over with optimized column design
- Save time with fast, user-friendly protocols
- No need to monitor pH or add isopropanol
- Buffers and columns available separately
- Significantly less plastic used when compared with other kits
- Responsibly-sourced and recyclable packaging
Monarch DNA Gel Extraction Kit Protocol
**Before you start, add 4 volumes of ethanol to 1 volume of DNA wash buffer.
**All centrifugation steps should be carried out at 16,000 x g (~13,000 RPM).
Excise Gel Fragment
Visualize your DNA fragment of interest under UV light and excise from the gel.
Tips: You have to cut the agarose as close to the DNA as possible and limit UV exposure to prevent DNA damage.
Dissolve Agarose
Transfer your excised band to 1.5ml microcentrifuge tube and weigh the gel. Add 4 volumes of Monarch Gel Dissolving buffer to the tube (eg, add 400μl of buffer to the 100μg of gels). Incubate your sample between 37 – 55°C for 5 – 10 mins to melt the agarose. Mixing periodically to ensure the agarose is completely dissolved.
Tips: Incomplete dissolving results in lower yield. This is because undissolved agarose still containing DNA and may also clog the column which reduces the elution efficiency.
Bind DNA to Column
Once the gel slice is dissolved, load your sample to the column. Centrifuge it for 1 min.
Wash the DNA
Reinsert the column into the collection tube and add 200μL of DNA Wash Buffer. Centrifuge for 1 min. Repeat the wash steps by adding 200μL of wash buffer and centrifuge for 1 min.
Elute DNA
Transfer the DNA into a clean microfuge tube. Add a minimum of 6μL of DNA elution buffer to the centre of the column. Incubate for 1 min. Centrifuge for 1 min and collect the flow through.
Tips: Make sure that the tip doesn’t contact the flow-through.
For video, please watch the Monarch DNA Gel Extraction Kit.
Tips for Optimizing DNA Quality and Yield
- Do not incubate gel slice above 60°C as this will damage the DNA.
- Add elution buffer to the centre of the column matrix for efficient elution.
- For fragments >8kb, add 1.5 volume water after dissolving agarose.
- For plasmids >10kb, heat the elution buffer to 50°C before use.
- Binding and elution spins can be reduced to 30 seconds.
Monarch RNA Cleanup Kit (10, 50 and 500µg)
You may need to purify your RNA before any application involves the use of purifying RNA. The Monarch RNA Cleanup Kit helps you to purify concentrated and high-quality RNA (>25nt) from enzymatic reactions like labelling, capping, in vitro transcription and DNase I treatment.
Like other purification kits, this kit purifies RNA through a bind/wash/elute steps, with the advantage of minimal incubation and spin times. The eluted RNA is ready for a wide variety of applications, such as RT-PCR, RNA labelling and RNA Library Prep for Next Generation Sequencing (NGS). If you need to purify the smaller RNA fragments, you can modify the protocol to do so.
There are three types of Monarch RNA Cleanup Kit, each of them comes with different binding capacity, 10ug, 50ug and 500ug.
Specification of Monarch RNA Cleanup Kit
| Description | Specification |
| RNA Sample Type | Cleanup and concentration of RNA from enzymatic reactions (labeling, capping, in vitro transcription reactions, DNase I treatment) |
| Binding Capacity | 10µg |
| RNA Size Range | ⥠25 nt ( ⥠15 nt with modified protocol) |
| Typical Recovery | 70–100% |
| Elution Volume | 6–20 µl |
| Purity | A260/280 > 1.8 and A260/230 > 1.8 |
| Protocol Time | 5 minutes of spin and incubation time |
| Compatible Downstream Application | RT-PCR, Small RNA library prep for NGS, RNA Library Prep for NGS |
Application of Monarch RNA Cleanup Kit
| Application | Explanation |
| RNA Cleanup and Concentration (including from the TRIzol aqueous phase) | RNA purified by other methods can be further purified |
| Enzymatic Reaction Cleanup | Enzymes such as RNA polymerases, DNase I, Proteinase K and phosphatases are removed allowing efficient desalting |
| In vitro Transcription Cleanup | Enzymes and excess NTPs are removed to yield highly pure synthesized RNA |
| RNA Gel Extraction | Purification of RNA from agarose gels |
| RNA Fractionation | Fractionation of RNA into small and large RNA pools |
Advantages of Using Monarch RNA Cleanup Kit
- Ideal for cleanup and concentration of RNA after enzymatic treatments including DNase I, Proteinase K, labelling, capping or in vitro transcription (IVT)
- Efficiently purify RNA ⥠25 nt (a simple modification enables purification of RNA ⥠15 nt)
- Elute in ⥠20 µl for concentrated RNA
- 70-100% RNA recovery
- Can be used to purify RNA from the aqueous phase following TRIzol® or similar extractions
- Simplified workflow with a single wash buffer
- Unique column design prevents buffer carryover and elution of silica particulates
- Columns and buffers are available separately
- Purified RNA is ready for use in a wide variety of downstream applications, including transfection
How to purify using Monarch RNA Cleanup Kit
** Add 4 volumes of ethanol to wash buffer
**All centrifugation steps should be carried out at 16,000 x g (~13,000 RPM).
Prepare your sample
Recommended starting sample volume: 50µL. If your sample volume is lower, you can adjust the volume with Nuclease-free Water. However, if your sample volume is large, you have to scale buffer volume accordingly.
Add binding buffer
Add 2 volumes of RNA Cleanup Binding Buffer to your sample.
For RNA ⥠25 nucleotides, add 1 volume of ethanol.
For RNA ⥠15 nucleotides, add 2 volumes of ethanol.
Tips: Do not VORTEX for mixing. Mix well by pipetting up and down or invert the tube gently.
Load sample onto the column
After you load your sample to the column, spin at 16000xg for 1 min and discard the flow through. If your sample more than 900µL, repeat the steps as necessary as the column can only hold 900µL at a time.
Wash column
Reinsert the column into the collection tube. Add 500µL RNA Cleanup Wash Buffer and spin for 1 min, then discard the flow-through.
Repeat the column wash
Add 500µL RNA Cleanup Wash Buffer and spin for 1 min, then discard the flow-through.
Elution
Transfer the column to RNase-free 1.5ml microcentrifuge tube. Be careful of the tips of the column do not contact with the flow-through to prevent the traces of salts and ethanol are not carried to the next steps. If the tips contact with the flow-through, respin for 1 min.
After centrifuging, add different volume of RNase-free water to the column and incubate for different incubation time according to the binding capacities of Monarch RNA Cleanup Kit in the table below.
| Binding capacities of Monarch RNA Cleanup Kits (µg) | Volume of RNase-free Water* (µL) | Incubation Time | Spin Time (min) |
| 10 | 6-20 | n/a | 1 |
| 50 | 20-100 | n/a | 1 |
| 500** | 50-100 | 5 mins (room temp) | 1 |
* Yield may slightly increase if a larger volume is used, but the RNA will be less concentrated.
** A second elution can be performed to maximize RNA recovery.
After the addition of RNase free water (and incubation for T2050), centrifuge to elute RNA. The eluted RNA can be used immediately or stored at -70°C.
For video, please watch Monarch RNA Cleanup Kits Protocol.
Tips for using Monarch RNA Cleanup Kits
- Before loading the sample to the column, be sure to add RNA Cleanup Binding Buffer first, then add ethanol. The binding buffer serves to inactivate enzymatic activities. Hence, preserving samples integrity. Addition of ethanol ensures that the DNA bind to the column.
- Do not add more than 900uL to the column at one time as this will exceed the column reservoir capacity.
- Ensure the tip of the column does not touch the collection tube after washing.
- During elution, load the RNAse free water to the centre of the column matrix.
- Recovery of small RNA, <45nt is affected by the degree of secondary structure. If the recovery of small RNA is lower than expected, you can dilute the samples with 2 volumes of ethanol, instead of 1 volumes, in the earlier steps of the protocol. The additional ethanol disrupts the secondary structure and enhances the binding to the column. As a result, the ethanol shifted the cutoff size of RNA from 25nt to 15nt.
- To save time, you can reduce the spin time for binding and elution to 30 seconds, instead of 1 min.
Monarch Genomic DNA Purification Kit
To study the genomic DNA from living things, we first need to extract and purify the DNA from the biological samples. In Monarch Genomic DNA purification kit, it contains a comprehensive solution for cell lysis, RNA removal, and purification of intact genomic DNA. This kit helps you to extract genomic DNA from a wide variety of samples, such as cultured cell, blood and mammalian tissues as well as tough to lyse sample, including bacteria and yeast. If you want to extract and purify gDNA from the clinically-relevant sample, you may consider Monarch Genomic DNA Purification kit as your option.
Purified gDNA using this kit has a high DIN score and minimal RNA contamination. And the best thing is, it is ready for any downstream application. For example, library preparation for NGS, PCR and qPCR. On top of that, it is an excellent choice for upstream of long-read sequencing platforms as the purified gDNA has a peak size of >50kb.
What you should know before using Monarch Genomic DNA Purification Kit
| Description | Specification |
| Input | Cultured mammalian cells: up to 5 x 106 cells Mammalian whole blood: 100 µl Tissue: up to 25 mg, depending on tissue type Bacteria: up to 2 x 109 Yeast: up to 5 x 107 Saliva: up to 500 µl Buccal swabs Genomic DNA requiring cleanup |
| Binding Capacity | 30µg genomic DNA |
| Genomic DNA Size | Peak size > 50 kb for most sample types; may be lower for saliva and buccal swabs |
| RNA Content | D< 1% (with included RNase A treatment) |
| Elution Volume | â¥35 µl, but 100 µl is recommended |
| Purity | A260/280 ⥠1.8 A260/230 ⥠2.0 |
Validated Sample Types
| Mouse | Tail Ear Liver Kidney Spleen |
Heart Lung Brain Muscle Blood |
| Rat | Liver Brain Muscle |
– |
| Others | Deer Muscle Human Blood Rabbit Blood Pig Blood Guinea Pig Blood Cow Blood Horse Blood Dog Blood Chicken Blood HeLa Cells |
HEK293 Cells NIH3T3 Cells E. coli Rhodobacter sp. B. cereus T. kodakarensis S. cerevisiae Saliva Buccal swab |
Advantages of using Monarch Genomic DNA Purification Kits
- For use with a broad range of sample types, such as cells, blood, and tissues (including fatty and fibrous tissues)
- Also works with tough to lyse samples (e.g., bacteria, yeast)
- Suitable for clinically-relevant samples (e.g., saliva, cheek)
- Obtain excellent yields of highly-pure DNA
- Extremely low residual RNA contamination (typically <1%)
- Isolates high molecular weight gDNA (peak size typically ⥠50 kb)
- Use DNA directly in downstream applications, including PCR, qPCR and NGS
- Fast, user-friendly protocols with short lysis times and minimal hands-on time
- Can also be used to clean up previously extracted gDNA
- Includes RNase A
- Kit components available separately
Monarch Genomic DNA Purification Kit Protocols
Basically, the Genomic DNA Purification consists of two stages:
- Sample lysis
- Genomic DNA Binding and Elution
The protocols would be different for different sample types. You can check the details protocols below:
- Cells
- Tissues
- Blood
- Gram Negative Bacteria
- Gram Positive Bacteria and Archae
- Yeast
- Buccal Swab
- Saliva
Tips for Optimal Results
Tips on optimizing gDNA yields and quality
- Place the column in the centrifuge in a consistent orientation to optimize yield and purity. This ensures that the liquid takes the same path in each spin.
- Preheat the elution buffer to 60°C. This helps to increase the efficiency of elution and increase the yield.
- If more concentrated DNA is desired, reduced the elution volume from 100 to 35uL. However, this will lead to a reduction of yield for about 20-25%.
- Apply your sample only to the membrane and avoid upper column area.
- Do not transfer foam that formed during lysis.
- Avoid touching the column reservoir with pipette tips.
- Bind your DNA at 2 speeds to maximize binding and purity, first at 1000g and latter at maximum speed. Spinning at the low speed gives DNA more opportunity to bind to the matrix and spinning at the maximum speed help to remove the remaining salts and proteins.
- Invert the column with wash buffer to optimize removal of salts.
- Confirm the heating block temperature is 60°C. If the temperature is above 60°C, the DNA may be partially denatured.
Tips for efficient lysis of tissue sample
- Use the recommended input amount of sample type according to the Monarch DNA Inputs, as using excess sample will clog the column and results in lower yield and poor DNA quality.
- Cut the tissue into small pieces to increase the surface area for enzymatic digestion.
- Ensure no tissue pieces are stuck at the bottom of the tube after lysis buffer and proteinase K are added.
- Carry out lysis in a thermal mixer to increase efficiency. If time is not limiting, extend the lysis time up to 3 more hours to obtain higher yields and lower RNA content.
- If using heat block, ensure to vortex sample every 20-30 mins to speed up lysis.
- To ensure optimal RNA digestion, add RNase A at the end of lysis to prevent degradation of the RNase A by the Proteinase K.
- When working with fibrous or fatty tissue, or tissues stored in RNAlater, centrifuge lysate or 3 mins at max speed. As if these fibres are not removed prior to adding to the column, they will stick to the membrane and prevent DNA binding, results in reducing yield and quality.
Conclusions
To sum up, NEB has a series of Monarch Nucleic Acid Purification kits. These Monarch kits enable quick and easy purification of high-quality DNA and RNA. The DNA and RNA purified from these kits are suitable for use in a variety of downstream applications. With fast, user-friendly protocols and optimized buffer systems, you can focus your time on the experiments that will drive your research forward!























One thought on “Nucleic Acid Purification Kits”