What are the Restriction Enzymes (RE)?
Restriction Enzymes (aka ‘molecular scissors’) are enzymes produced by bacteria to cut the specific site of DNA, which also known as the recognition site or restriction site. Each restriction enzyme can recognize these unique recognition sites of DNA and cut the DNA into fragments.
Do all RE recognize and cut at the same restriction site?
However, not all REs recognize and cut at the same restriction site.
Type I REs cleave DNA randomly that far from their recognition site. Though they are of noteworthy biochemical interest, they have little practical value as they do not produce discrete DNA fragments or distinct gel-banding patterns.
On the other hand, type II REs cleave DNA at a specific site, which often underlying close to or within the recognition sequence. They do not cut randomly and often result in discrete DNA fragments or distinct gel-banding patterns. Hence, researchers usually use type II REs in the research for molecular cloning, DNA mapping, gene sequencing, etc.
What happens after the restriction enzymes cut the DNA strands?
Restriction enzymes are sometimes known as restriction endonucleases because they often cut within the DNA molecule. This cutting results in the formation of either sticky ends or blunt ends of DNA, depending on the restriction endonuclease you use. The sticky ends and blunt ends are important for the ligation, which will be explained later.
Sticky Ends and Blunt Ends
What are Sticky Ends?
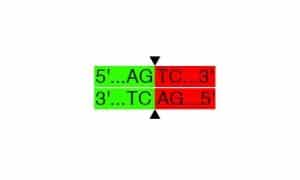
Sticky ends are known as cohesive/non-blunt ends. The REs (EcoRI-hf and BamHI-hf) cut the DNA strands a few base pair from each other. This cutting results in the formation of 5′- or 3′- of unpaired DNA strands. We name them as 5′- (5 prime) or 3′ (3 prime) overhang.
Most of the time, the sticky ends are palindromic in sequence. The sequence of 5′ to 3′ on one strand is the same as the 5′ to 3′ on the other strand.

Cutting site of EcoRI. 5′-overhang is created after the DNA strands are cut. You can see the palindromic sequence at the sticky end, 5′-AATT-3′.
What are Blunt Ends?
Blunt ends refer to the base pair on both strands of a DNA end at the same base pair. We also name blunt ends as non-cohesive ends, since there is no unpaired DNA strand fleeting at the end of DNA. Example of the REs that create the blunt ends is AfeI and AluI.
Cohesive-end Ligation and Blunt-end Ligation
After the restriction enzymes cleave the DNA strands, DNA ligase acts as a catalyst to join the phosphate backbone covalently.
In the cohesive-end ligation of DNA and plasmid, the overhangs that cut from the same RE are complementary to each other. Due to the attraction of hydrogen bondings from both overhang strands, they can ‘stick’ easily with each other. This stabilization of hydrogen bondings increase the efficiency of DNA ligation and thus, require less DNA and plasmid when compare to blunt-end ligation.
In blunt-end cloning, although the DNA and plasmid do not benefit from the attraction of hydrogen bonding, they can still be ligated in the presence of DNA ligase. The advantage of using blunt ends in DNA ligation is they are universally compatible with other blunt-ended DNA. However, the yield of blunt-end ligation is generally lower as a lack of complementary overhangs, in which the ends of DNA are not ‘stick’ together. The absence of this hydrogen bonding decreases the efficiency of ligation since a successful ligation depends on the transient association of 5′-phosphate and 3′-hydroxyl groups of DNA as well as DNA ligase.
Restriction Enzymes in LabMal
Nearly 300 REs from New England Biolabs is available in LabMal! NEB is indeed the leader in RE technologies for over 40 years. Until now, NEB dedicates to providing the best quality, the highest purity and unparalleled performance.
Below shows the features of RE from NEB:
- More than 215 enzymes are active in a single buffer – CutSmart™ Buffer.
- More than 195 enzymes are Time-saver qualified, meaning you can digest 5-15 mins, or digest DNA safely overnight.
- High-Fidelity (HF®) restriction enzymes are the engineered enzymes with reduced star activity.
- Each vial of restriction enzyme comes with a 6X purple loading dye.
If you do not know what REs you should use, you must try the NEBcutter V2.0! You can easily find the REs that you need to cut the DNA sequence.




I hope that you won’t stop writing such interesting articles. I’m waiting for more of your content. It’s so good that i’m going follow you.